Dr. Carolyn Bertozzi and her team at Lawrence Berkeley laboratory are using molecular self-organization tendencies to give cells the orderly neighborhoods they desire. However, this microscopic community has an unusual dress code-- the cells display DNA on the outside of their membranes, which allows them to keep each other in line.
 Unlike the cells' nuclear DNA, the exterior DNA doesn't serve a genetic purpose. It is a purely structural construct. Last week I talked about how self-assembly can form data storage surfaces (and islands of Cheerios). Systems with small, mobile parts seek out orientations that are stable and ordered because these require the least energy.
Unlike the cells' nuclear DNA, the exterior DNA doesn't serve a genetic purpose. It is a purely structural construct. Last week I talked about how self-assembly can form data storage surfaces (and islands of Cheerios). Systems with small, mobile parts seek out orientations that are stable and ordered because these require the least energy. Self-assembly is also responsible for forming proteins and DNA, which have far more structural complexity than a geometric arrangement of cereal. It isn't difficult to self-assemble large, complicated structures from small, simple ones-- all it takes is a choosier joint. For example, this milimeter-scale self-assembled LED device from the Whitesides group at MIT involves 14-sided self-assembled blocks; the specific shape of the solder on large faces provides a "key" for specific alignment.
Though the blocks can brush against each other in any way in a solution, the shape of the solder pattern encourages a specific low-energy order.
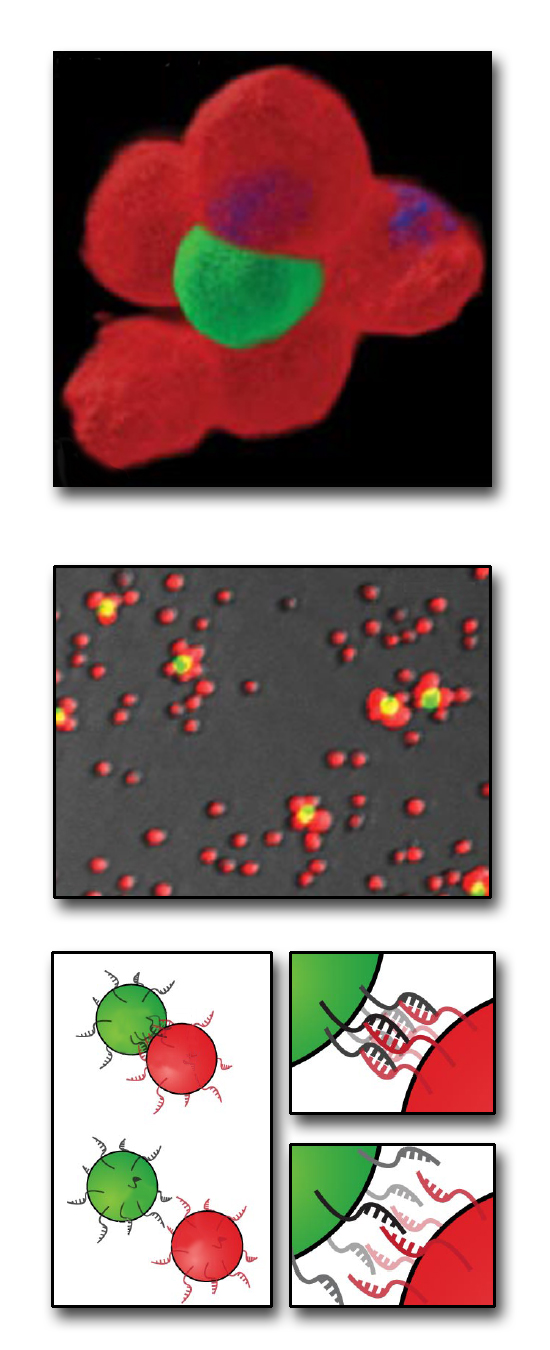 To get this kind of mechanical specificity on the surface of the cell, researchers Gartner and Bertozzi turned to another biological construct that self-assembles, DNA. DNA's signature two-strand helical structure is held in place by binding between a sequence of nucleotides on one strand to complementary nucleotides on the other strand. Each of the nucleotides in DNA will only form strong attachments to its complementary nucleotide: adenine only binds to thymine, and guanine only to cytosine.
To get this kind of mechanical specificity on the surface of the cell, researchers Gartner and Bertozzi turned to another biological construct that self-assembles, DNA. DNA's signature two-strand helical structure is held in place by binding between a sequence of nucleotides on one strand to complementary nucleotides on the other strand. Each of the nucleotides in DNA will only form strong attachments to its complementary nucleotide: adenine only binds to thymine, and guanine only to cytosine. The lonely nucleotides on a single strand of DNA will attract their complementary nucleotides in a solution, spontaneously recreating the second strand of the DNA double-helix. This also works for entire complementary strands of DNA-- they'll stick together like two puzzle pieces. By attaching strands of DNA to the outside of the cells, the researchers can design complementary cell surfaces which keep designated cells together like two best friends holding hands.
Creating a large object from smaller ones is something we take for granted at human scale. We build houses from wood and bricks rather than carving them out of a giant slab or pouring cement into a four-story mold. It's harder to do that with microscopic building materials; human-sized hands can't pick up cells and put them in order. To build on the microscale, we generally rely on top-down techniques-- we refine smaller features from large objects much like Michelangelo chiseled the David.
Like Dr. Frankenstein before them, Gartner and Bertozzi made their microscopic monster from smaller parts: a bottom-up assembly technique. This mimics nature's additive approach and gives the Frankentissues functions that more haphazard human-built tissues lack. For example, Gartner and Bertozzi combined blood progenitor cells with a hamster ovary cell that expressed a necessary growth factor for the progenitor cell's growth.
When the cells were simply mixed, the progenitor cells didn't grow. When the cells were assembled into a designer tissue, the progenitor cells communicated, formed a network, and grew.
Though the authors suggest using it to provide better seeding for tissue scaffolding, it's likely to have great applications in the field of thrombogenicity and biocompatibility of permanent implants, as well.
Gartner, Z. J. and Bertozzi, C. R. "Programmed assembly of 3-dimensional microtissues with defined cellular connectivity."
Jacobs, H. O. et al. "Fabrication of a Functional Cylindrical Display using Solder-Based Self-Assembly." Science 296, 323-325 (2002).



Comments