Physics can explain the cycle of the earth around the sun, but what drives cellular cycles? Two of the most important cycles in cells are the series of events that take place when cells divide, and circadian rhythms - the cycle of day-night events that even bacteria participate in.
Unlike a planet, a cell's cycle can't be described by simple physical laws, nor does it have a CPU like a computer to keep time and and control the order of events. So how do cells control their cycles? Researchers have been trying to reverse engineer cell cycles, and two recent successes give a fascinating molecular view of what's going on.
Your bike chain drives your wheels forward, but when you stop pedaling, your wheels still spin. It turns out, a yeast cell does the same thing. In order to replicate itself, a cell executes a series of complex events, each of which has to be precisely timed: the cell has to make sure it has the resources to divide ready, it then has to replicate its DNA, and finally it has to actually divide - splitting all of the contents of the cell in two, making sure each new cell gets all of the right chromosomes. Obviously, if any of these events happens out of order, the result is a major problem. If a cell tries to divide before copying its DNA, there won't be enough DNA to go around. If a cell copies it's DNA more than once before dividing, the new cells will be loaded with extra chromosomes, which also creates problems. So how does a cell control the timing of these events?
The cell's equivalent of a bike chain is something called a cyclin/CDK complex, which is a pair of proteins that orchestrate the major events of cell division. But just which events are controlled by this pair has been unclear, especially when it comes to regulating genes during the cell division process. Before we look more closely at the cell's cyclin/CDK bike chain, we need to take a detour and look at how genes behave when cells divide.
Obviously, cells don't keep all of their genes on all of the time. During cell division, genes are often turned on when they're needed, and off when they've done their job. In a yeast cell, about 1000 genes get turned on and off at some point during the cycle of cell division, as you can see below in this figure showing the behavior of about 1000 genes during the cell cycle:
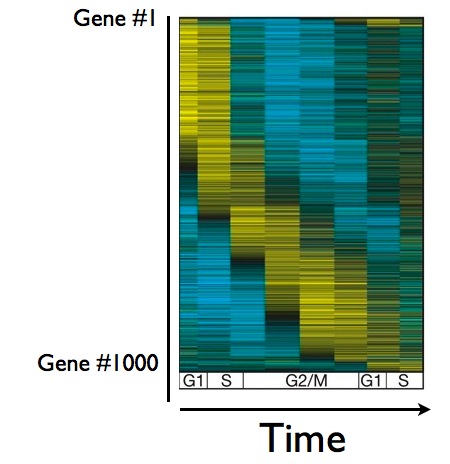
Here's what the figure means: each row corresponds to the behavior of one gene, and as you move from left to right, you can follow the behavior of that gene over time. Yellow means the gene is on, blue means it's off. As you can see, a group of genes comes on early in the cell cycle (the blob of yellow in the upper left corner), other genes come on later - you get the picture. The question is now, how much of this gene behavior controlled by the cyclin/CDK master timer, and how much of it is a separate, self-perpetuating cycle? In other words, if you stop pedaling during cell division, does the wheel of gene regulation still spin?
A group led by Steven Haase at Duke University demonstrated that yes, it does keep spinning. They shut down the cyclin/CDK complex, and followed the behavior of these cell cycle genes. Without the driving force of the cyclin/CDK complex, the cells themselves froze up - they didn't replicate their DNA, and they didn't divide. But about 70% of their cell cycle genes kept oscillating in a self perpetuating cycle:
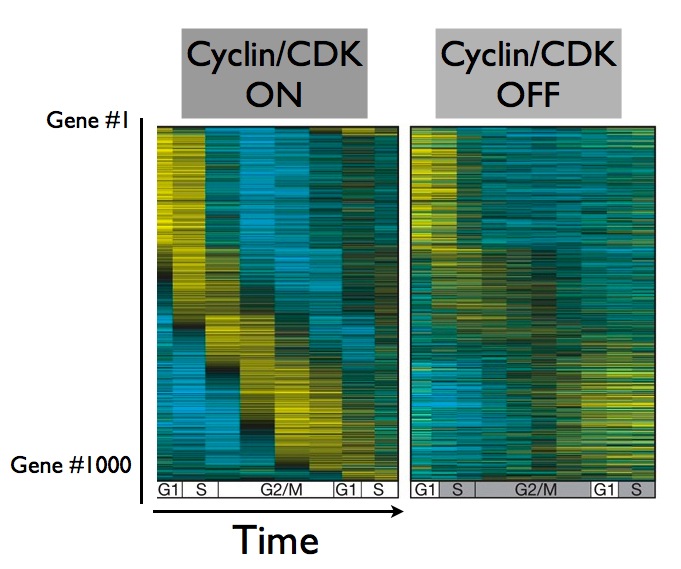
What this teaches us is that cell division is actually made up of at least two cycles - one driven by the cyclin/CDK protein complex, and another self-perpetuating cycle of gene expression. And when it comes to cell division, what goes for a yeast cell usually goes for a human cell as well - scientists believe it's critical to understand how the cell division cycle is driven, because cell division plays such a crucial role in so many biological process, from the development of a single stem cell into groups of specialized cells like muscle or nerve cells, to diseases like cancer.
In addition to the cell division cycle, cells have a cycle that follows a major rhythms that dominates everything on our planet - the cycle of day and night, which produces what's called a circadian rhythm. You feel the effects of your circadian rhythm when it's out of whack, after a long transatlantic flight, or after staying up all night drinking beer and watching South Park reruns. Amazingly, single-celled organisms like cyanobacteria have a circadian rhythm as well.

Unlike the cycle of gene expression in yeast, the cyanobacterium circadian clock doesn't need to have sets of genes turned on and off. In fact, it doesn't even need most of the bacterium: you can take just three key proteins from Synechococcus elongatus, put them in a test tube, and they will spontaneously start cycling.
What exactly is cycling? One of these key three proteins, called KaiC, gets modified in a rhythmic fashion: it gets decorated with phosphate molecules, and once it's fully decorated, the phosphates are pulled off. Once the phosphates are off, they get put back on again - in cycle after cycle. Inside the actual bacterium, this cycle drives important biological events, but in a test tube, these three proteins, including KaiC, just participate in a perpetual cycle of dressing and undressing KaiC. What happens is this (follow the black line, which tells you how dressed or undressed KaiC is by phosphates)
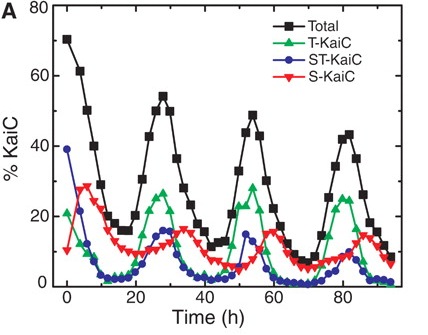
Here's the key question: How does KaiC know whether it's getting dressed or undressed? When it's halfway up or down in a cycle, how does it know whether the cycle is going up or down? Obviously this 3-protein circus is able to come together in a test tube and put on this cycling act; how does it work? A group lead by Erin O'Shea from Harvard University figured out the answer, which turns out to be fairly simple, although it took a lot of biochemistry to sort it out.
It turns out that there is an order to how KaiC gets dressed and undressed with phosphates, and that order only happens when all three necessary proteins are placed in a tube. Phosphates can be attached at two different types of amino acids in the KaiC protein - either at a serine or a threonine. In the cyanobacterium cycle, phosphates get put on threonines first, and then on serines. When it comes time to pull the phosphates off, they go in the same order: off the threonines first, and then off the serines. So that's how this cycle knows whether it's coming or going. If you look closely at the figure above, you can see the details: T-KaiC (that is, KaiC with phosphates only on threonine - the green line) shows up early in the cycle, while S-KaiC (KaiC with phosphates only on serine - the red line) is the last form of KaiC to go down.
Why should you care? Besides the amazing fact that such simple biological devices can be made by tossing three proteins in a test tube, it also shows us how reverse engineering these small, simple systems that we can really start to gain insight into how biological parts form coherent wholes. Three rocks in a tube don't oscillate, but proteins, by sheer chemistry, can. Biochemical cycles drive so much of our biology, so learning how to reverse engineer these cycles, starting in microbes, is important science.



Comments