HOUSTON - (April 18, 2016) - Thank the little "muscles" in your neurons for allowing you to remember where you live, what your friends and family look like and a lot more.
New research at Rice University suggests actin filaments that control the shape of neuron cells may also be the key to the molecular machinery that forms and stores long-term memories.
The Rice lab of theoretical biological physicist Peter Wolynes reported in the Proceedings of the National Academy of Sciences a theory about how long-term memories are made; the theory is based on simulations that analyze the energy landscapes of the proteins involved.
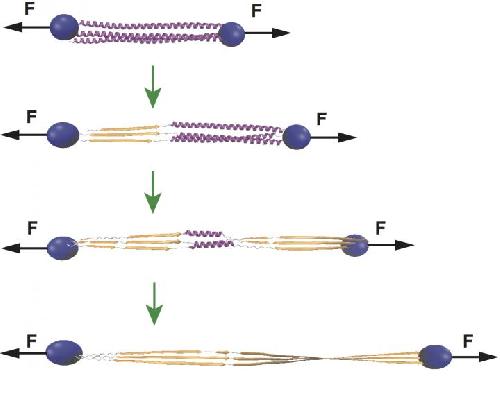
Wolynes and his colleagues are pioneers in the development of an energy landscape theory for proteins, which enables them to build computer models of proteins to predict how they will fold. These molecular-dynamics simulations employ the principle of minimal frustration by which proteins find their most stable folded forms.
For long-term memories, stability is desirable. Wolynes and his co-authors, Rice graduate student Mingchen Chen and postdoctoral researcher Weihua Zheng, determined the path to encoding memories may lie in the way actin filaments - the "muscle" part of the cytoskeleton in every eukaryotic cell - pull upon and stabilize soluble cytoplasmic polyadenylation element binding proteins (CPEB) into longer, insoluble prion-like fibers.
Prions are proteins that, when they misfold, are thought to become self-propagating and cause infectious diseases like mad cow disease, Creutzfeldt-Jakob disease and other disorders. But their very existence and the transitions known to take place in synapses suggest properly folded prions must have a biological function, the researchers wrote. These transitions were the focus of their study.
CPEB proteins, when made in cells, first bind a few at a time in oligomers, which are coiled alpha helices. The intrinsic energy landscapes of these oligomers allow mechanical forces provided by actin to prompt a transition into longer beta strands that are much more stable. These now-stable fibers are thought to aggregate and encode memories in neurons' synaptic regions.
Wolynes said Francis Crick, co-discoverer of the structure of DNA, was onto something 20 years ago when he wrote about memory and molecular turnover. Crick puzzled over the fact that memories tend to last much longer than proteins typically do in living cells. "Crick slightly anticipates, in one sentence, that perhaps what we have is a form of protein that aggregates somewhere," he said. "By virtue of being an aggregate, it's not able to move. In that way it would be able to mark one particular synapse.
"It's obviously very difficult to study the molecular basis of memory because memory involves a fairly complex activity," Wolynes said. "You can't study it in a bacterium. You have to study it in some sort of organism that can learn.
"At the same time, it's clear that forming memories involves some very high-order neural processing and other things at the subcellular level in order to store the large amount of information you memorize. There are many steps in memory that are really not understood at all."
He said previous research shows that memories make changes in the synapse, the thousands of regions in each neuron responsible for sending electrical and chemical signals to other neurons. "Short-term memories that last less than an hour or so seem to be done through the electrical and direct biochemical circuitry. Forming these memories doesn't seem to require creating new protein," Wolynes said.
Researchers who conducted experiments with sea slugs poisoned to prevent them from synthesizing proteins seemed to confirm that, he said. "They found these snails were able to memorize things for short periods of time but not for periods of hours when protein synthesis was stopped."
Chen, who led the Rice research, knew from the literature that actin has the ability to bind oligomeric CPEB. This fact, along with the computer simulation, suggests that the mechanical force provided by actin can restructure CPEB into a longer fiber with new hydrogen bonds between the coils. Wolynes said that the restructuring not only forces CPEB to a lower-energy, prion-like state, but also allows the prion to bind an RNA sequence that otherwise prevents more actin from being synthesized. The resulting feedback loop further stabilizes the memory.
"We still don't understand the beginning of the process, how you go from short-term to long-term memory," he said. "But we can now see that actin starts to form in a particular location in response to electrical signals. The actin then takes any CPEB oligomers that are around and activates them, which makes more actin and causes the formation of a self-replicating prion of the CPEB. That prion aggregates until it stops, changing the structure of the synapse in a way that should last for a very long period of time, perhaps decades."
Wolynes said he put Chen on the job with few expectations. "I give starting students a project I think will teach them the tools we use to look at protein dynamics," he said. "It's usually a somewhat far-outish project, and if they don't get anywhere, I won't feel sad.
"So I said, 'Why don't we look at this protein that (Eric) Kandel and (Susan) Linquist said was involved in memory, this CPEB protein.'"
Wolynes said many neurobiologists have followed up on that pioneering work with sea slugs. "But we add a new element by being able to look at the structures of these proteins and to predict the thermodynamics of the process," he said. "We now can see how the force of the cytoskeleton can complete a feedback loop that allows the memories to be preserved."
Wolynes considers the new study a beachhead to launch others to determine the entire process of how memories form, as well as the implications for diseases like Alzheimer's and Parkinson's that involve protein aggregation.





Comments